Prediction of side effects
Novel compounds often fail during development due to non-tolerable side effects. Binding assays deliver data on off-targets and indicate putative side effects, however do not verify effects in a complex arrangement of cells. Therefore, compounds might be withdrawn from further development without examining their real physiological effects. Hence, there is a need for predictive tests uncovering side effects early during preclinical development.
Our in vitro side effect assay allows predicting either pro-convulsive/seizurogenic effects or sedative effects. The analysis is performed based on concentration/response data and our multi-parametric data analysis and allows screening test compound libraries very early in drug discovery. The calibration is based the functional differentiation of
(1) known sedative drugs vs. activity-reducing drugs
(2) known pro-convulsive vs. activity-increasing compounds.
Prediction of sedative properties
Many compounds show inhibition of general activity parameters such as spike rate. However, these parameters alone are not sufficient enough for prediction. Our classification technology uses the multitude of parameters and was calibrated using clinically-relevant data from used drugs with known sedative effects and those without sedative effects but still activity-decreasing.
Known strong sedative drugs: Diazepam, Flunitrazepam, Propfol, Memantine and Methaqualone.
Non-and little-sedative drugs: Amisulpride, Aripiprazole, Flumazenil, Fluoxetine, Fluvoxamine.
The classification successfully distinguishes the two groups and predicts the sedative properties of many known clinical hypnotic drugs but also those with unwanted side effects.
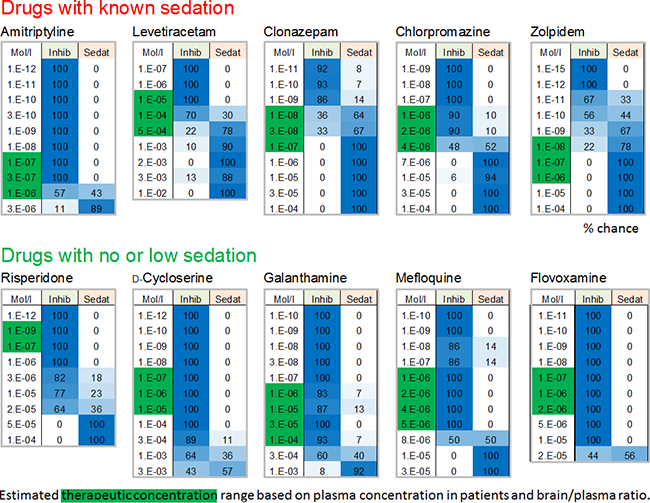
Image: Analysis of internal reference datasets from our clinical database showing whether and if, at which concentrations, the drugs exhibit a chance (values in %) to be sedative or in comparison are only inhibitory (non-sedative). Drugs with known sedative properties (either major effect or side effects) were correctly classified as sedative within their therapeutically-relevant concentration range (green box). Those, which lack sedative side effects exhibit low % values for sedation. The estimated clinical/therapeutic concentrations were calculated based on literature data on plasma concentration and brain/plasma ratio from humans.
Prediction of pro-convulsive/seizurogenic properties
Many compounds show excitation of general activity parameters such as spike rate. However, these parameters alone are not sufficient enough for prediction of seizurogenic neurotoxicity. Our classification technology uses the multitude of parameters and was calibrated using clinically-relevant data from used drugs with known seizurogenic effects and those without seizurogenic effects but still activity-increasing.
Known seizure compounds: Picrotoxin, Pentylenetetrazole, Bicuculline.
Excitatory compounds: NMDA, Orexin A, Mefloquine
The classification successfully distinguishes the two groups and predicts the sedative properties of many known clinical hypnotic drugs but also those with unwanted side For the classifier calibration 3 excitatory and 3 state-of-the-art pro-convulsive compounds were selected. All compounds show increase of spike rate. Selected concentrations (boxes) were used for further analysis. The aim was to establish a classifier based on excitatory data points which is able to sort the respective compounds into their correct category.
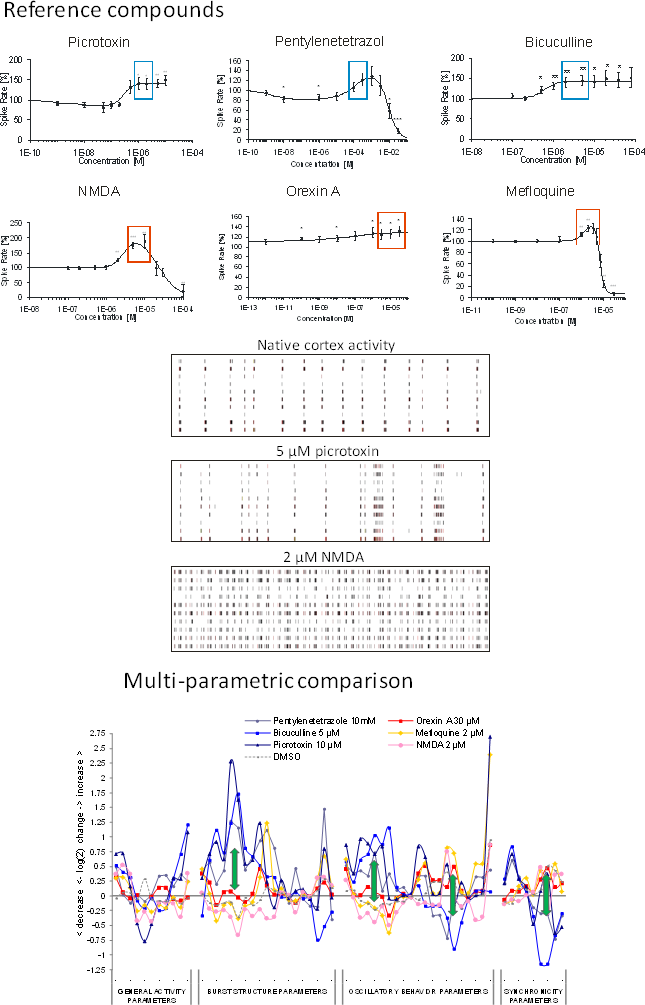
Image: reference datasets from our substance database. Compounds with seizurogenic and excitatory/non-seizurogenic compounds increase activity (boxes) but exhibit completely different spike train pattern (center). Right: Multi-parametric data analysis of selected concentrations shows opposite effects at burst structure, regularity and synchronicity parameters (arrows).
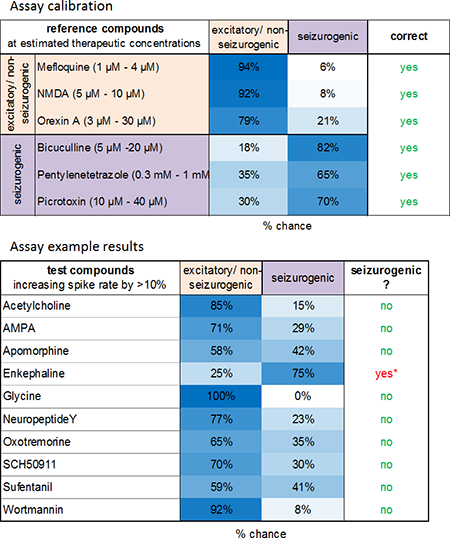
Image: The classification successfully distinguishes the two groups (left). Prediction of seizurogenic/pro-convulsive properties of compounds at concentrations which increase spike rate >10 % revealed pro-convulsive effects of enkephaline also shown by Clynen et al. 2014, Mol. Neurobio. 50(2).
 infobox@neuroproof.com
infobox@neuroproof.com +49 381 54345-660
+49 381 54345-660
